Metabolomics Core Laboratory
Metabolomics Core Laboratory
 |
 |
|
PI: Prof. Ming-Shi Shiao Research Expertise: Chemical biology, lipid metabolism, metabolomics |
Assoc. Prof. Mei-Ling Cheng Research Expertise: Metabolism, Free Radical Biomedical Science and Translational Medicine
|
Researchers:
Prof. Ming-Shi Shiao and Assoc. Prof. Mei-Ling Cheng
Conceptual Framework:
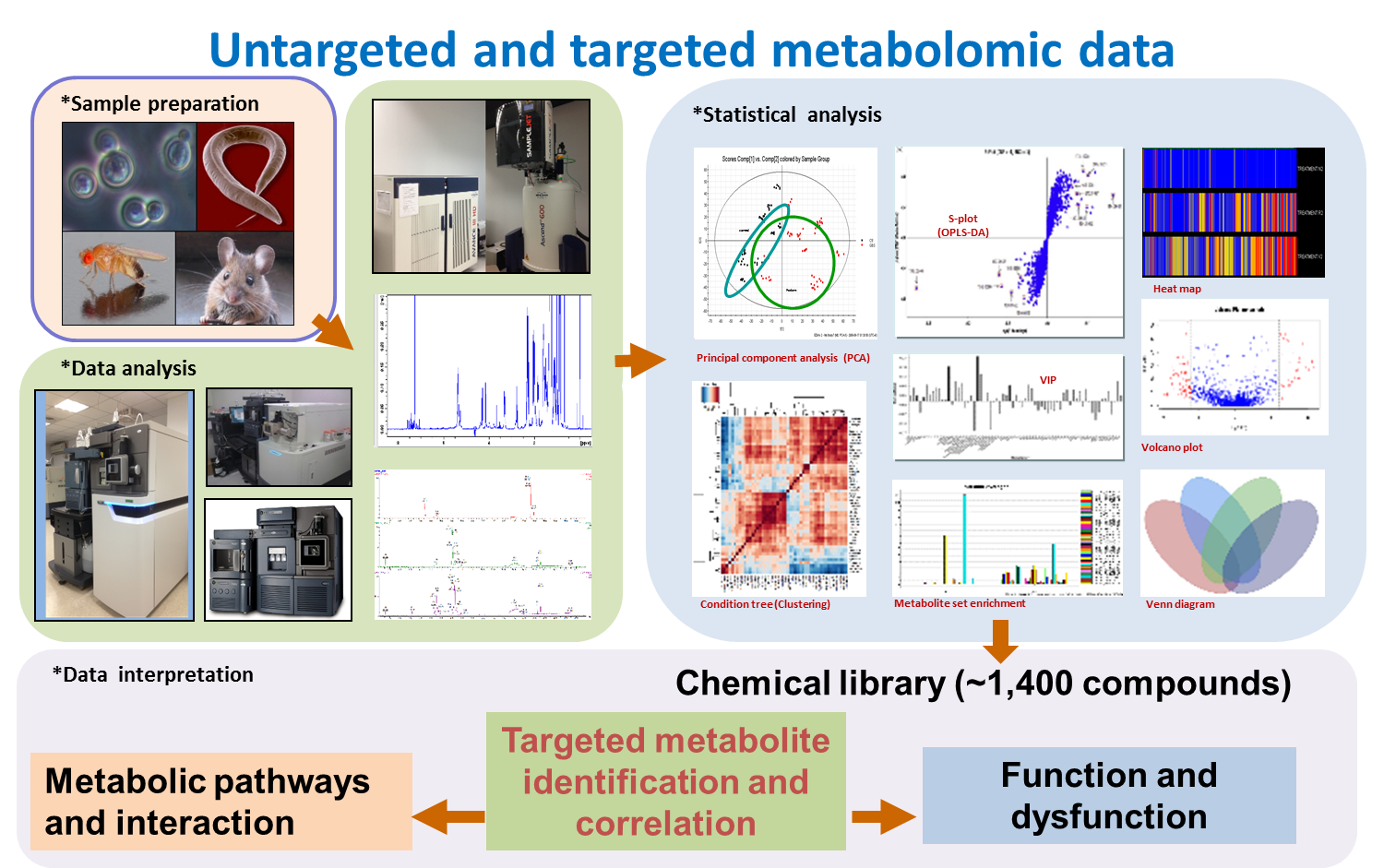
General description:
The mission of the core Lab is to provide the research platform, key technology, and standard operating protocols for metabolomics studies.
Background:
Metabolomics is a newly emerging field concerned with the comprehensive characterization of low-molecular-weight (LMW) metabolites in biological systems. Information obtained from metabolomics approach may provide a global view of the metabolic pathways, network, and their interactions in normal and pathophysiological states.
Objectives:
Accordingly, metabolomics-based information may help to detect disease progression, monitor response to treatment, and identify new targets for drug design. Since a global, unbiased approach is undertaken, metabolomics is most applicable to human degenerative diseases including diabetes, cardiovascular disease, and neurodegenerative diseases, which involve the interactions of multiple risk factors in disease pathogenesis and progression.
Methods:
Metabolomics approach relies on nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) as the key tools for untargeted metabolite profiling and target metabolite identification. Plasma, urine, CSF, cells and biospecimens are samples for analysis. Precise determination of metabolite concentration is not always necessary in an early stage. If specific metabolites (target metabolites) turn out to be important, the information of their concentrations becomes instructive.
Publications:
1. Huang JP, Cheng ML, Wang CH, Shiao MS, Chen JK, Hung LM. (2016) High-fructose and high-fat feeding correspondingly lead to the development of lysoPC-associated apoptotic cardiomyopathy and adrenergic signaling-related cardiac hypertrophy. International Journal of Cardiology 215:65-76.
2. Cheng ML, Chang KH, Wu YR, Chen CM. (2016) Metabolic disturbances in plasma as biomarkers for Huntington's disease. Journal of Nutritional Biochemistry 31:38-44.
3. Chiu CY, Lin G, Cheng ML, Chiang MH, Tsai MH, Lai SH, Wong KS, Hsieh SY. (2016) Metabolomic Profiling of Infectious Parapneumonic Effusions Reveals Biomarkers for Guiding Management of Children with Streptococcus pneumoniae Pneumonia. Scientific Reports 6:24930.
4. Martel J, Wu CY, Hung CY, Wong TY, Cheng AJ, Cheng ML, Shiao MS, Young JD. (2016) Fatty acids and small organic compounds bind to mineralo-organic nanoparticles derived from human body fluids as revealed by metabolomic analysis. Nanoscale 8(10):5537-45.
5. Chiu CY, Yeh KW, Lin G, Chiang MH, Yang SC, Chao WJ, Yao TC, Tsai MH, Hua MC, Liao SL, Lai SH, Cheng ML#, Huang JL. (2016) Metabolomics Reveals Dynamic Metabolic Changes Associated with Age in Early Childhood. PLoS ONE 11(2): e0149823.
6. Wang PY, Lin WC, Tsai YC, Cheng ML, Lin YH, Tseng SH, Chakraborty A, Pai LM. (2015) Regulation of CTP Synthase Filament Formation During DNA Endoreplication in Drosophila. Genetics 201(4):1511-23.
7. Wu YH, Chiu DT, Lin HR, Tang HY, Cheng ML, Ho HY (2015) Glucose-6-Phosphate Dehydrogenase Enhances Antiviral Response through Downregulation of NADPH Sensor HSCARG and Upregulation of NF-κB Signaling. Viruses 7(12), 6689-6706.
Contact: 03-2118800 #3650
